CIDER is a webserver being developed by the Pappu Lab for calculating parameters relating to disordered protein sequences. Specifically, this will calculate various parameters which help translate primary sequence information into better understanding how the unstructured ensemble might behave, as well as annotating your sequence on the diagram of states (from ref [1]), which is shown below.
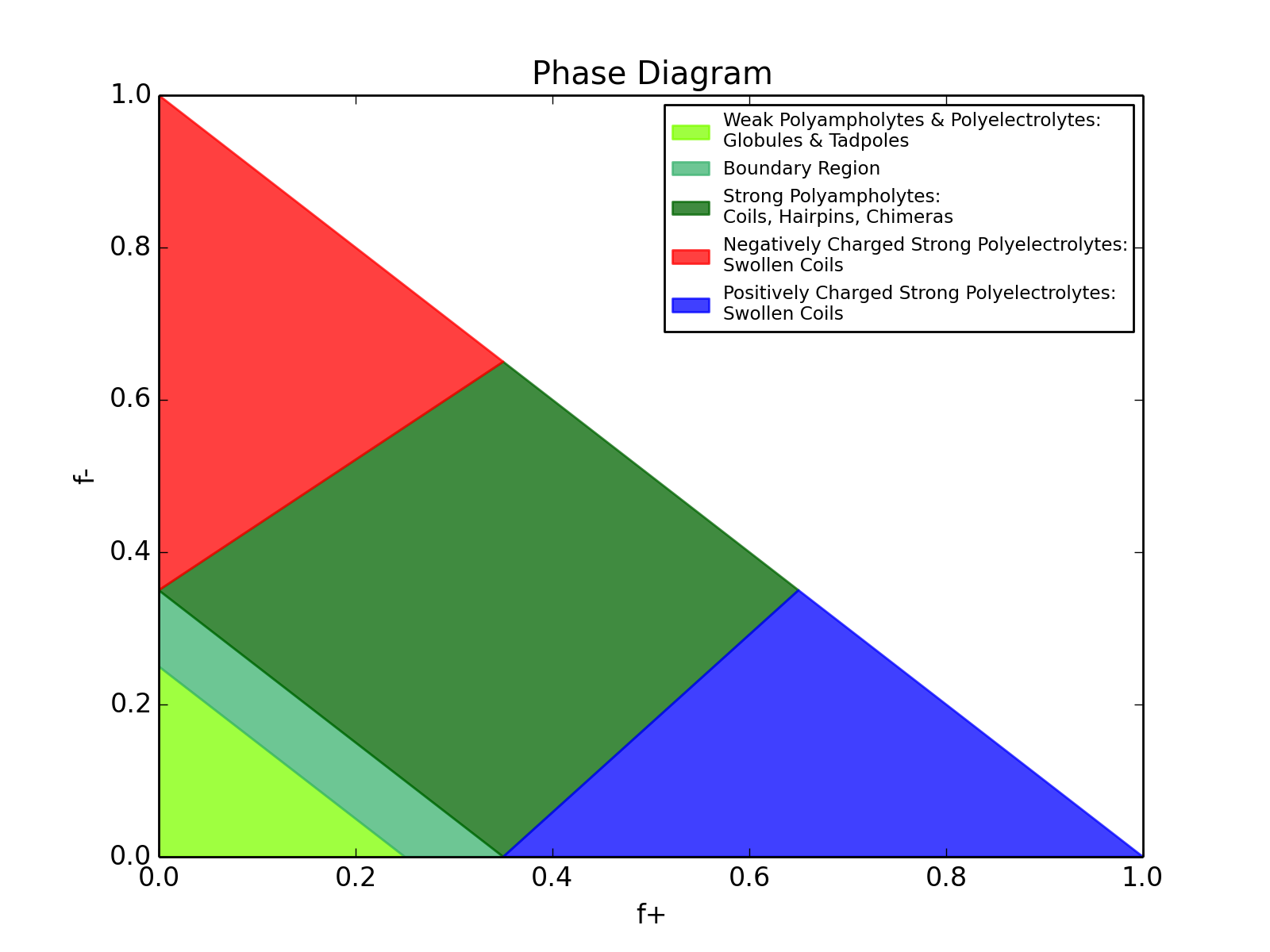

CIDER was developed by Alex Holehouse, based on code written by James Ahad. For any questions please contact Alex at alex.holehouse@wustl.edu.
Please cite CIDER and localCIDER as;
A.S. Holehouse, R.K. Das, J.N. Ahad, M.O.G. Richardson, R.V. Pappu (2017) CIDER: Resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophysical Journal, 112: 16-21 Access Article
Also, please make sure to cite the original papers were specific sequence insights were first determined (see the CIDER paper for an appropriate list).
[1] Das & Pappu (2013) Proceedings of the National Academy of Sciences USA. 110: 13392-13397